Why Work At Pierian?
Flexibility
The decisions you will make as part of our team will impact the lives of people around the world. We enable our team to do their best work by putting a healthy work/life balance front and center.
Growth
You will join us in an exciting field where new knowledge and breakthroughs happen every day. Our culture encourages our team members to be in the driver’s seat of their own careers and embrace new opportunities.
Diversity and Inclusion
We pride ourselves on hiring and nurturing a diverse workforce. Not only do we want to make the world a more equitable place, but we know that people who feel accepted and valued are more fulfilled and empowered to reach their full potential.
Who Are We?
Across all our teams and departments we hold ourselves to high values, learn about them here.



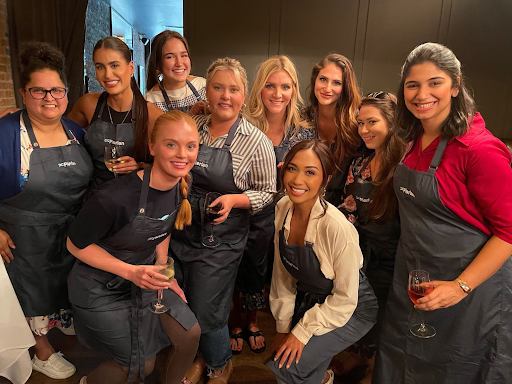

Teamwork

Teamwork
- We hold each other accountable and drive each other to excel
- We value collaboration, respect, inclusivity, and diversity in thought
- We always assume positive intent, and if unsure, we seek to clarify
Passion

Passion
-
We go above and beyond for each patient, customer, and person
-
We drive results because we are inspired by what we do
-
We challenge the norm and encourage out-of-the-box thinking
Excellence

Excellence
-
We perform at the highest levels and deliver the best-in-class products
-
We consistently innovate and improve
-
We continue to learn and support employee growth
Integrity
Integrity
-
We do the right thing because it is the right thing to do
-
We operate honestly and believe in being true to yourself
-
We do what we say we are going to do
Compassion

Compassion
-
We believe the how matters in getting results
-
We strive to make the world a better place
-
We treat others as they would like to be treated
Employee Benefits

Dental, Vision, and Pet Insurance
401(k) Plan with Matching
Holiday Pay & Sick Leave
Life & Supplemental Life Insurance
Disability & Health Insurance
"Even after 7 years in the company, the people amaze me every day. I have become a leader myself by following in the footsteps of the leaders here. It's a great company to work with, with the smartest people in it, and a founder who will keep on inspiring you every day."
People here are curious about their work and do it with dedication, I love the collaborative work people do here, it keeps me motivated.
More importantly, the leaders and my manager like to play a positive-sum game, unlike the industry where people cut each other in order to progress.
The work environment is positive and healthy. The employee compensations are great and company policies really cater to the needs of all employees. The colleagues are extremely professional and dedicated to what they do and the management is always looking for ways to make things easier for the employees. Great place to learn and grow!



